Our Interests
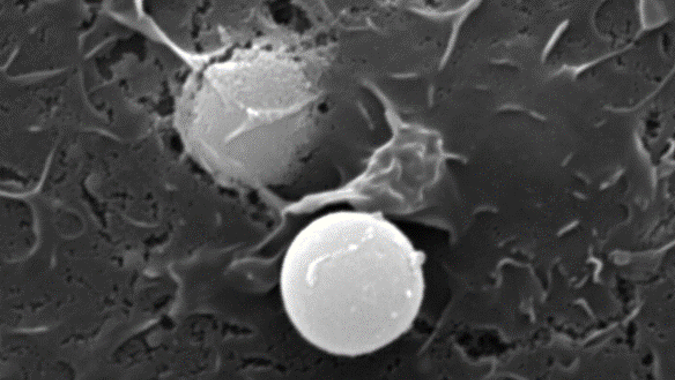
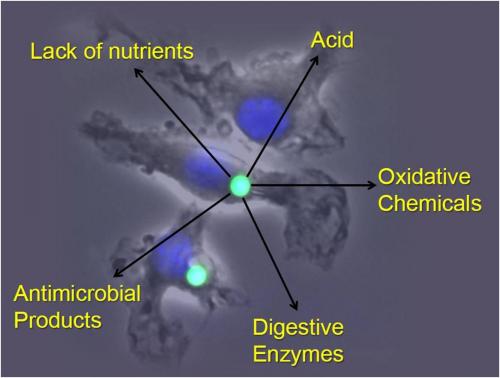
Phagocytes are cells such as macrophages and dendritic cells that can engulf material via phagocytosis. They function in a diverse array of essential tasks throughout the body, operating at the frontlines to protect the body against invading microbes and also performing numerous housekeeping functions that are necessary for normal growth and maintenance of health. The phagosome is the organelle that is formed within the phagocyte following phagocytosis and is charged with the task of processing the engulfed material appropriately. In primitive phagocytes, such as the amoeba, this organelle serves as the “stomach” of the cell and acts to efficiently and completely digest its contents for cellular nutrition. In phagocytes of higher order organisms, however, the phagosome can be charged to perform a multitude of precise functions, many of which rely on controlled or partial deconstruction of engulfed material. Phagocytes such as macrophages and dendritic cells can adapt the lumenal chemistries within the phagosome to best suit the task at hand. This allows these cells to perform numerous tasks within the body. However, with increased versatility comes increased fallibility which can lead to disease.
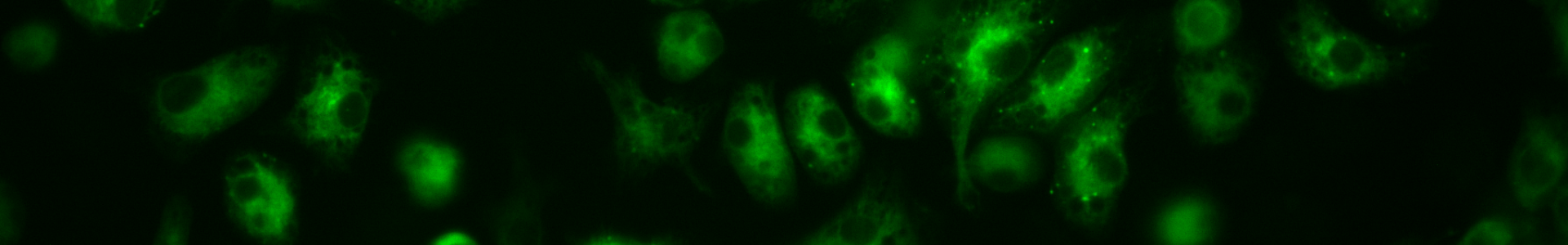
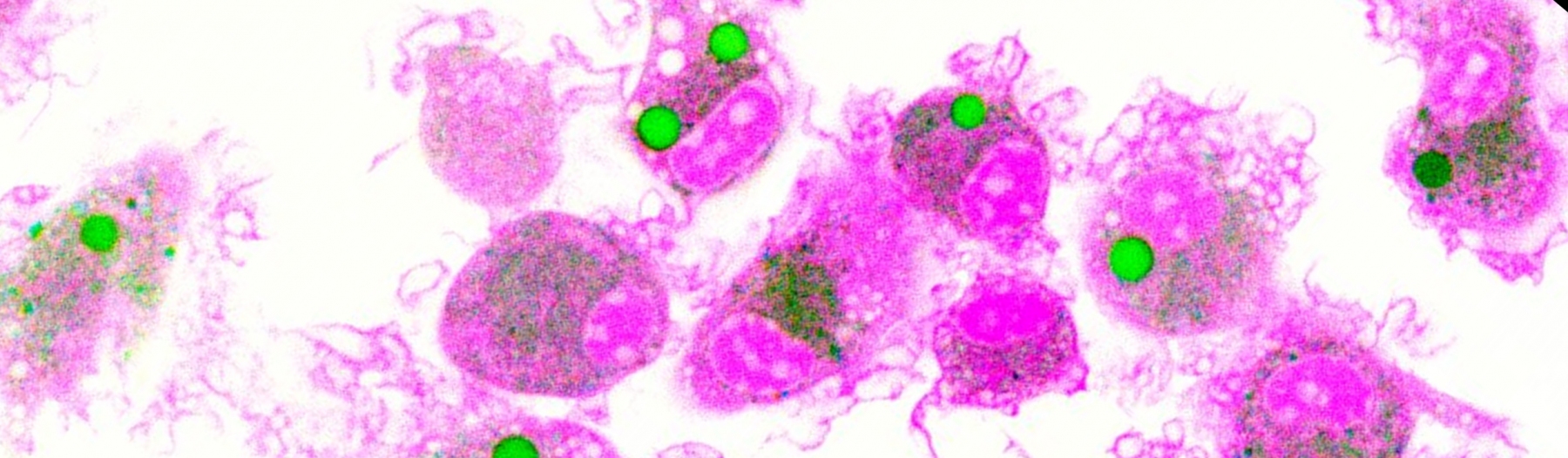
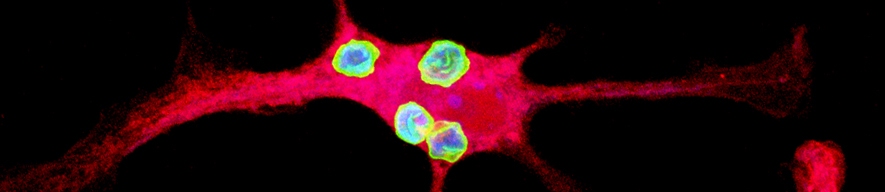
The Yates lab focuses its research on the lumenal microenvironment within phagosomes in macrophages and dendritic cells (pictured below). By utilizing techniques in fluorometry, microscopy and molecular biology the group can monitor chemistries within this microenvironment; explore relationships between phagosomal chemistries and dissect pathways that alter phagosomal function. Since phagosomes play key roles in numerous physiologies and pathologies, this fundamental work has the potential to impact a diverse array of biomedical fields from infectious disease to autoimmunity.
Our Projects
Eructophagy – Phagocytes Burping Out Their Meals
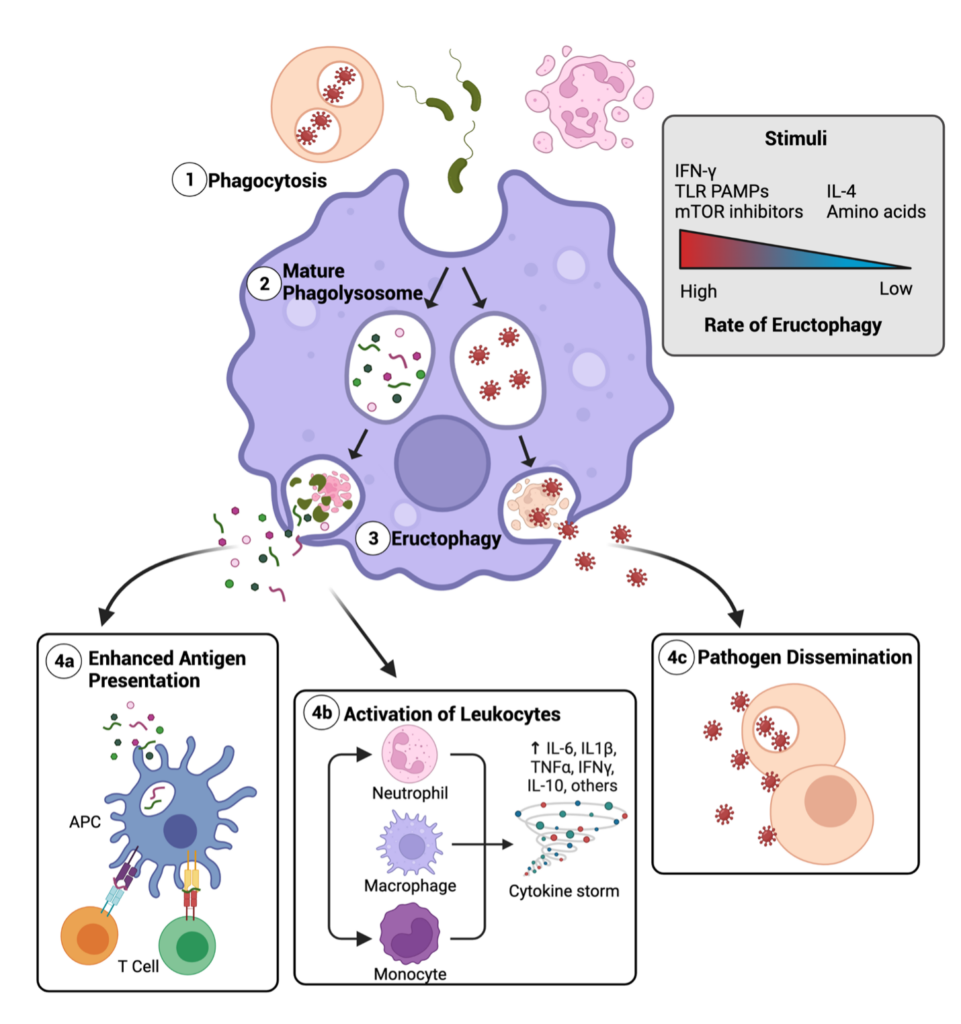
Eructophagy is a novel phagocytic event where the phagolysosome fuses with the cell membrane to release soluble matter to the extracellular space. “Eructophagy” is derived from eructare (Latin: to burp or belch), and the suffix -phagy to acknowledge the connection with phagocytosis and autophagy. To date, we have determined that eructophagy is induced by proinflammatory stimuli, is negatively regulated by mTOR, and is dependent on key autophagy proteins (Greene et al. 2022). We propose that eructophagy releases PAMPs and DAMPs to amplify local inflammation, with the potential to release pathogenic factors, hydrolases and peptides in the phagolysosome. Hence, the impact of eructophagy in antigen presentation, pathogen dissemination, tissue damage and overall health and disease is widespread.
Phagosome Biochemistry in Professional Phagocytes
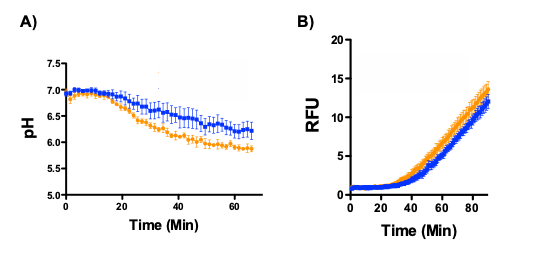
How does the redox environment, proteolytic activity and acidity of the phagosomal lumen impact a phagocyte’s ability to process and present antigen? To maintain tissue homeostasis? To ameliorate pathogens? Utilizing several kinetic assays that employ sophisticated fluorescence microplate readers (including EnVision, Fluorostar Optima, FlexStation and the InCell 2000) with lab-developed, optimized bead-reporters, the research objectives of the Yates Lab revolve around the complicated biochemical relationships inside the phagosome microenvironment.
Antigen Presentation
The Yates Lab possesses several transgenic mouse strains to study physiological interactions between antigen-presenting cells of innate immunity and T cells of the adaptive immune system and T cell biology as well as in models of multiple sclerosis. We utilize peptide pulse-chase assays, flow cytometry and fluorometric assays to study TCR-ligand interactions, T cell activation and cross-presentation of antigens.
Cathepsin Z and the Inflammasome
Cathepsins are lysosomal proteases that are acquired by the phagosome after phagolysosome biogenesis. Cathepsins function in protein degradation and antigen presentation. Of the various phagolysosomal cathepsins, we demonstrated that cathepsin Z (CatZ) is associated with an increased incidence of multiple sclerosis in a mouse EAE model (Euan et al. 2017). Furthermore, decreasing CatZ expression was associated with reduced EAE clinical score and IL-1B levels, thus presenting a novel role and mechanism of action for a cathepsin protease. We are interested in understanding how the cysteine CatZ (which is also an exopeptidase) impacts the generation IL-1B, a cytokine released upon induction of the NLRP3 inflammasome.
GILT
Proteolysis in phagolysosomes via cysteine cathepsins (B, L S, Z and K) is optimized under acidic conditions. The active site cysteine must be reduced by thiol reductases in order to be functionally active, but is sensitive to oxidation during the phagosomal generation of reactive oxygen species. It is known that phagosomal disulfide reduction is catalyzed by the thiol-dependent enzyme GILT (gamma-interferon inducible lysosomal thioreductase). It was determined that GILT is essential for disulfide bond reduction for protease function in the digestion of phagolysosomal cargo and antigen processing for presentation.
TRPM8/A1 and Icilin
The supercooling drug, icilin, and its primary receptor TRPM8 (transient receptor potential melastatin-8) have been described by our lab to attenuate autoimmune inflammation in a murine model of multiple sclerosis by modulating the T-cell response (Ewanchuk et al. 2018). However, icilin-mediated activation of TRPM8 also induces an undesirable off-target cooling effect that precludes this compound as a potential therapeutic for T cell-driven neuroinflammation. Thus, our goal is to screen candidate drugs that can activate TRPM8 or TRPA1 without off-target cooling.
BADASS Project
How are peptides being cleaved by proteases in the phagolysosome? The BADASS (Bead Assisted Discovery of Antigen Scissile Sites) project aims to profile digestion patterns of common or intriguing antigens generated in the phagolysosome. Antigenic proteins of interest are covalently attached to experimental particles that are then engulfed by antigen-presenting cells (eg. macrophages and dendritic cells) under various experimental conditions. The beads and remaining protein fragments are recovered and analyzed to determine the differences in fragment repertoires between experimental conditions.
Our Techniques
Fluorescent Microscopy
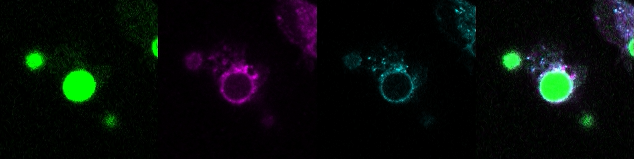
Measuring Biochemistries in the Phagosome Microenvironment
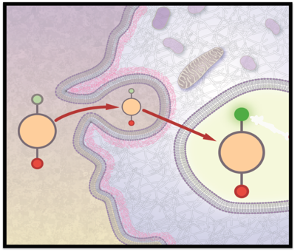
FRET (Förster resonance energy transfer or fluorescence resonance energy transfer)

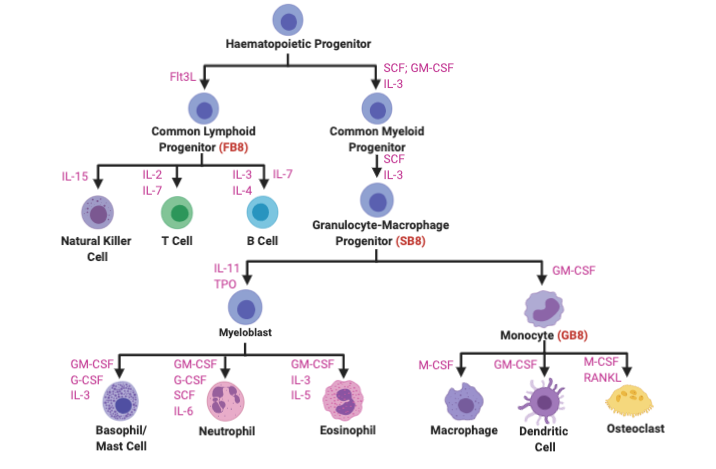
HoxB8 Conditionally Immortalized Cells
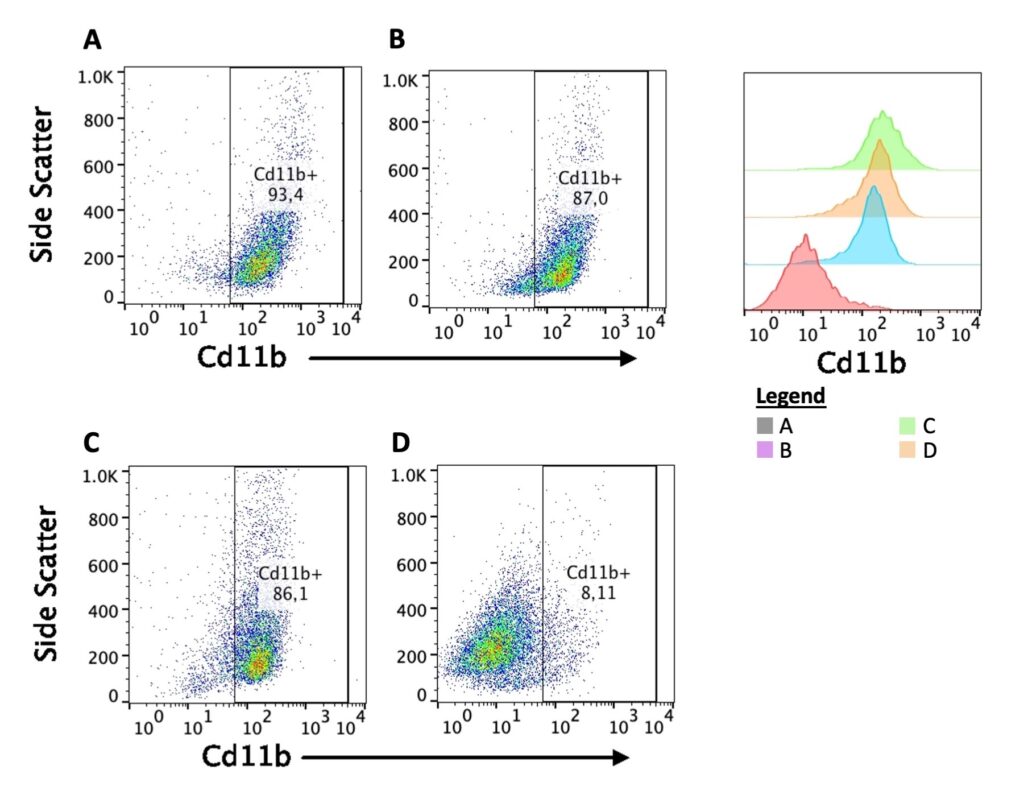
Flow Cytometry
Acidification, Proteolysis and Redox Assays