Graduate Student and Postdoctoral Fellow Projects related to identifying the microbial actors and pathways, chemical species, and MIC mechanisms that lead to facility failures
Investigating MIC Risk in Offshore Oil Recovery Operations
Danika Nicoletti, University of Calgary
Supervisor Dr. Lisa Gieg, University of Calgary
My research examines the risk of microbiologically-influenced corrosion (MIC) in offshore produced water samples from Floating, Production, Storage and Offloading (FPSO) vessels using microcosm experiments. Produced water is incubated with a carbon steel corrosion coupon in microcosms under anaerobic conditions and monitored over time for various chemical changes. Upon experiment termination, the planktonic and sessile microbial communities and the corrosion coupons are analyzed along with chemical data to investigate whether MIC was a factor in the degradation of carbon steel compared to sterile controls. An additional research project involves isolation of a sulfide-specific enzyme for later use in the development of a sulfide biosensor (Assays & Devices).
Danika completed her Master of Science thesis in 2020. Read her thesis.
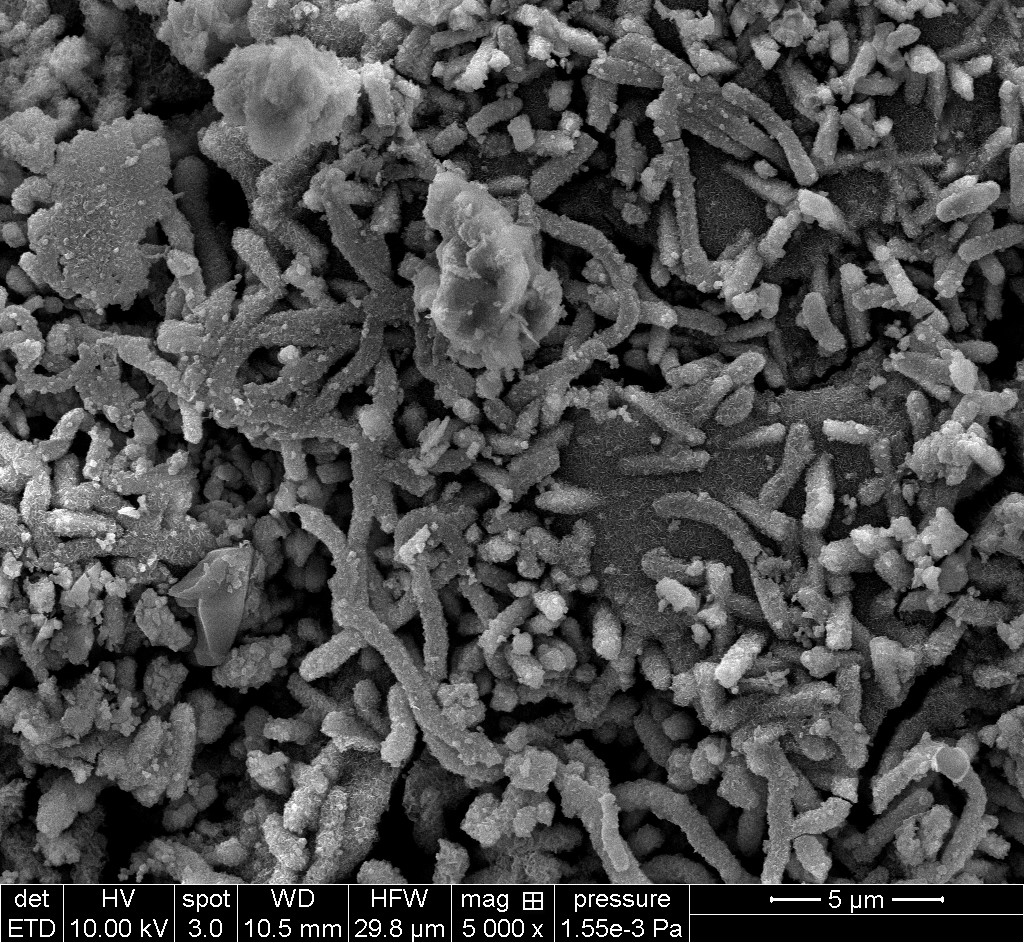
A scanning electron microscopic image of a biofilm on the surface of a corrosion coupon. Photo Credit: Danika Nicoletti, University of Calgary
Tracking multidrug resistance efflux pumps as a source of biocide tolerance in corrosion communities
Damon Brown, University of Calgary
Supervisor: Raymond Turner, University of Calgary
I am investigating the presence and abundance of multidrug resistance efflux pumps (MDREPs) in Oil and Gas environments as a mechanism for increased microbial tolerance towards commonly used biocides (or antiseptics) in this industry. MDREPs are frequently located on mobile genetic elements which allows these genes to move throughout a community in response to a biocide challenge, thus increasing the tolerance of the entire community towards the biocide after repeated use. Effectively tracking and quantifying these genes in a community can increase our ability to advise effective biocide treatments for operators of these systems, improving asset lifetime and safety.
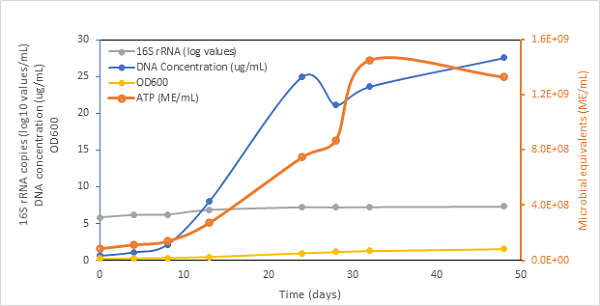
Growth curve of Acetobacterium woodii using field relevant techniques, ATP activity microbial equivalents/mL), DNA concentration (µg/mL), OD600 and copies of 16S rRNA (log10 copies/mL). Figure Credit: Damon Brown, University of Calgary
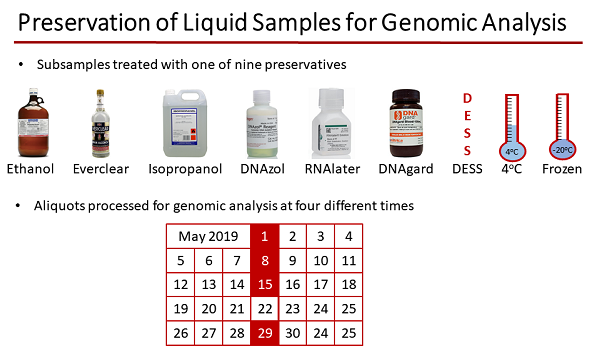
Experimental design for testing preservatives for their effectiveness in preserving genomic material in onshore process water samples. Image credit: Natalie Rachel & Nuno Fragoso, University of Calgary.
Preserving Microbial Community Integrity in Produced Water
Natalie Rachel, University of Calgary.
Supervisor: Lisa Gieg, University of Calgary
The nature a sampling location often determines their microbial signature, and these properties can vary wildly; in the event that the sampling material has a low concentration of biomass, large quantities will have to be collected. This is certainly the case when investigating the microbiology of oilfield systems, where produced water (PW) is one of the most common sources for sampling. As this field continues to develop, the importance for standardized operating protocols cannot be understated, so that industry can make the most informed operating decisions possible. Starting from the beginning with sample collection, it is imperative that their integrity be preserved as best as possible. To this end, we meticulously investigated the effectiveness of nine different preservation conditions on PW collected from the same sampling location within a heavy-oil producing field, and monitored how the community changed.
Investigating MIC Metabolism
Natalie Rachel, University of Calgary.
Supervisor: Lisa Gieg, University of Calgary
The vast biological diversity that exists amongst microorganisms complicates the creation of broadly applicable MIC detection strategies, as not all organisms are created equally: the severity of corrosion risk posed by microorganisms varies significantly depending on their biochemistry. While current approaches that involve the surveying of microbial populations are helpful in assessing MIC potential, these are indirect methods as the mere presence of microorganisms such as SRM does not necessarily indicate that corrosion will occur. In light of this, I am using genomics approaches to more concretely identify metabolic processes responsible for MIC; enzymes that are specifically responsible for oxidizing metallic iron may serve both as genetic diagnostic markers, as well as inhibition targets.

Internal pit associated with MIC found within a subsurface offshore pipeline. Photo Credit: Nuno Fragoso, University of Calgary
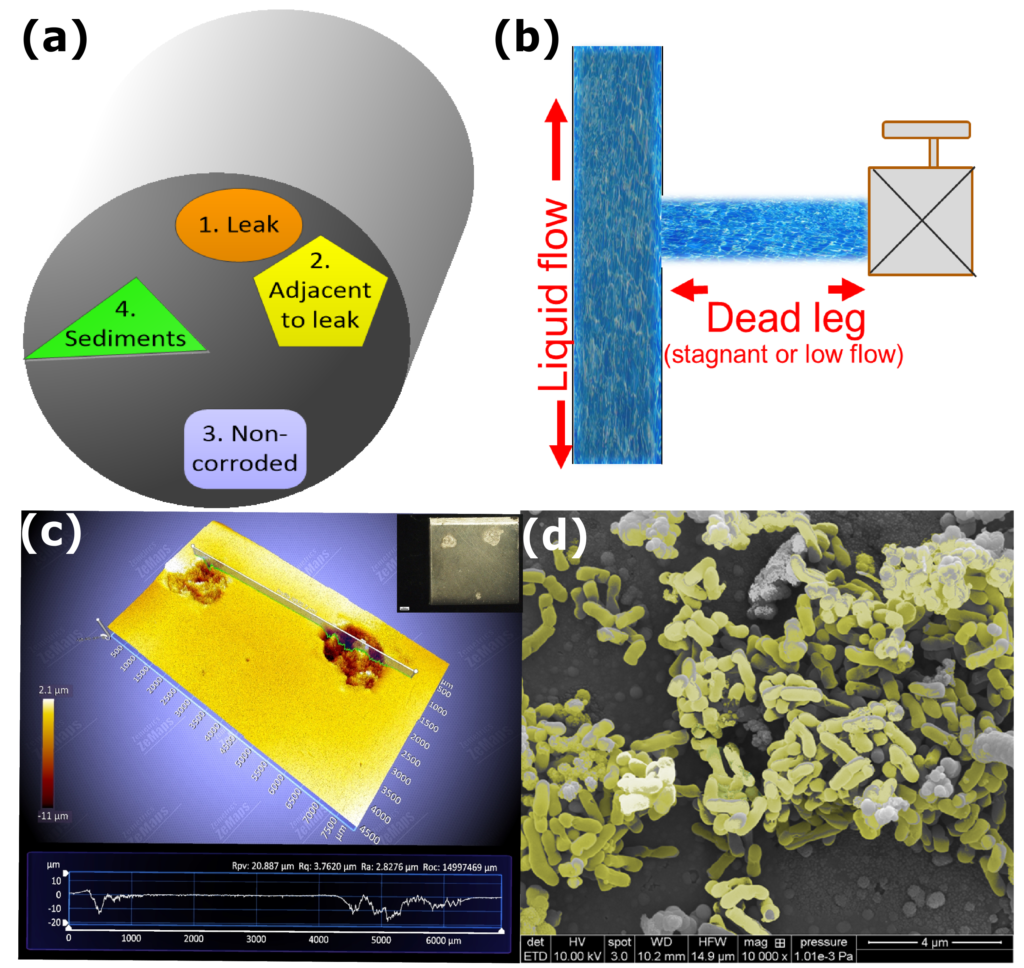
Schematic of pipe extracted from a bypass segment that experienced leak and the type of samples that were collected to perform a holistic MIC study. (b) Schematic of a bypass segment/deadleg area where fluids get accumulated due to stagnant or low flow conditions, increasing the probability of MIC/under deposit corrosion. (c) Profilometry image of a corrosion coupon after one month of incubation for MIC studies, the inset shows the pits visible on the surface of CS coupon (d) SEM image of a corrosion coupon immediately after corrosion assay where the biofilm attached on the surface of coupon is marked in yellow color.
Strategies for mitigating MIC
Mohita Sharma, University of Calgary
Supervisor: Lisa Gieg, University of Calgary
I am involved in developing mitigation strategies to prevent MIC and studying the potential role of biocides, biodispersants and corrosion inhibitors in controlling souring and corrosion in oil and gas operations. This involves:
- Conducting field visits for collection of environmental samples and subsequent assessment for their physical, chemical properties; microscopic analysis, study the microbial community present in such samples by next generation sequencing methods.
- Determination of corrosion rates by electrochemical and weight loss methods, sample evaluation using light and confocal microscopy, SEM, EDX, XRD and profilometry.
- Applying genomic and metagenomic tools in order to gain insight into community structure, activity and function of mixed culture communities especially bioelectroactive microorganisms in an engineered environmental system and natural environments.
Investigation of Internal Corrosion by Biological Activated Oilfield Deposits in Transmission Pipelines
Yessica Peralta, University of Alberta
Supervisor: John Wolodko, University of Alberta
My research investigates the corrosion rate of carbon steel in a packed vertical dead leg flow cell system simulating under deposit corrosion (UDC) and microbial induced corrosion (MIC) – biological activated oilfield deposits under anaerobically biotic conditions. Experiments search correlating real-time corrosion rate with the associated microbiological, chemical essays, and oily sludge composition (organic and inorganic) by employing electrochemical techniques. Identify the predominant mechanisms involved in both general and localized corrosion of carbon steel by interpreting and analyzing the monitored parameters (e.g. pH), electrochemical readings, coupon weight loss measurement, coupon surface morphology analysis, the chemical composition of corrosion products and sessile microbial community analysis. The Gutenberg editor uses blocks to create all types of content, replacing a half-dozen ways of customizing WordPress and aligning with open web initiatives.
Image of the Biological Activated Oilfield Deposit (BAD) – Under Deposit Corrosion (UDC) and Microbial Induced Corrosion (MIC).
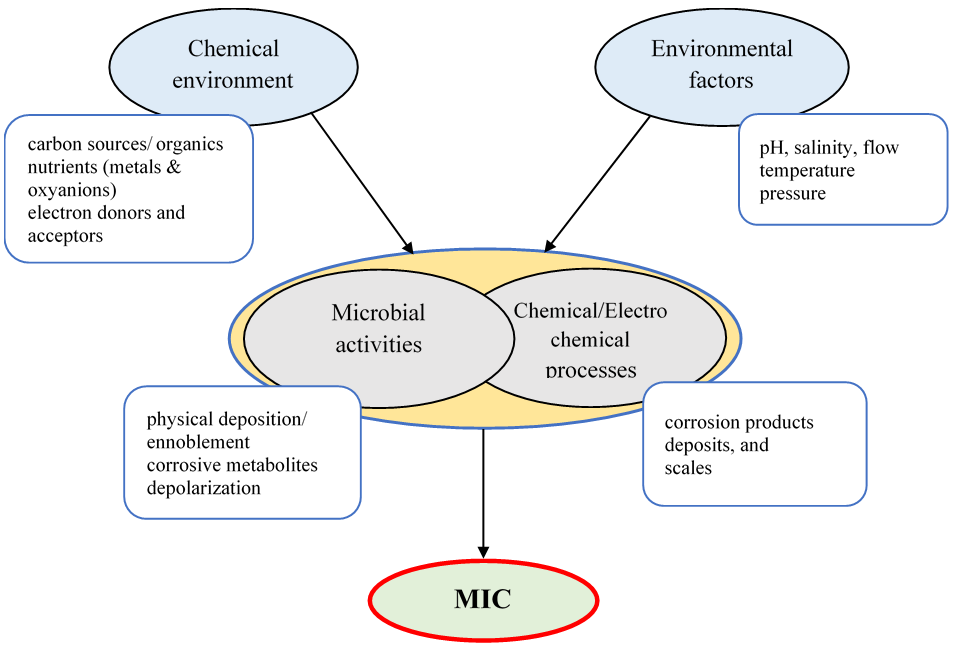
An outline of the interactions between chemical environment, environmental factors, electrochemical, and microbial processes
The effect of abiotic transformation of S and N species on MIC in offshore facilities
Abdulhaqq Ibrahim, Memorial University of Newfoundland
Supervisor: Kelly A. Hawboldt, Memorial University of Newfoundland
The chemical environment (e.g. produced water (PW), soured oil, limited oxygen environments) plays an important role on microbial activities leading to microbiologically influenced corrosion (MIC). One of the major causes of corrosion in the oil and gas facilities is H2S and/or other forms of sulfur via chemical transformations. Nitrogen compounds (nitrate/nitrite) present can also impact the chemistry of the system and microbial activities. In this research work, the key chemical species associated with MIC, resulting from biotic and abiotic processes were identified and analyzed considering the interactions between chemical environment, environmental factors, and microbial processes to understand the contributory effects on MIC. The sulfide and nitrite environment were studied as a function of temperature, pressure, nitrite level, and oxygen using equilibrium, and kinetic simulation approaches. We further conducted wet lab experiments with the field-sampled PW to investigate the reactivity of specific chemical species including sulfide and nitrite, generate kinetic data for key chemical reactions and validate the computer simulations. The work is anticipated to present a better understanding of the S and N chemistry in sulfide–containing chemical environment which can provide insight into the chemical–microbial relationships in oil and gas operations under different operating conditions.
Studying chemistry of MIC
Mahsan Basafa, Memorial University of Newfoundland
Supervisors: Kelly A. Hawboldt & Christina Bottaro, Memorial University of Newfoundland
Various analytical methods such as Ion-Chromatography, UV-vis Spectrophotometry, Gas Chromatography equipped with flame-ionization detection (GC-FID), and Gas Chromatography with sulfur chemiluminescence detection (GC-SCD) are being used at Memorial University for studying the chemistry of the environment leading to offshore MIC. Several organic electron donors typically encountered in oil field produced water can promote the activity of microorganisms leading to MIC. Due to low solubility, these compounds are usually found in trace quantities in aqueous phase, and pre-concentration is required before analysis. Furthermore, offshore produced water is a complex matrix containing various organic and inorganic compounds. Therefore, a selective sorbent material is needed to selectively extract and detect the target analyte(s). Thin-film molecularly imprinted polymers (MIPs) are engineered and fabricated in this study to selectively extract target organic compounds from produced water.

